Quickstart
Contents
Quickstart#
This tutorial provides a quick overview about the different tools available in evomap.
In general, input data is expected in the form of either higher-dimensional feature vectors, or in the form of pairwise relationships.
Given such data, evomap provides a flexible set of tools to process and manipulate the data, map it to lower-dimensional space, and to evaluate and explore the resultant maps.
Background#
Last updated: September 2023
This quickstart guide is based on the following paper. If you use this package or parts of its code, please cite our work.
References
[1]Matthe, M., Ringel, D. M., Skiera, B. (2022), "Mapping Market Structure Evolution", Marketing Science, forthcoming.
Read the full paper here (open access): https://doi.org/10.1287/mksc.2022.1385
Contact: For questions or feedback, please get in touch.
Module Overview#
evomap entails the following main modules:
evomap.preprocessing: Tools for preprocessing input data.evomap.mapping: Tools for mapping input data to lower-dimensional space.evomap.printer: Tools for drawing and annotating maps.evomap.metrics: Tools for evaluating maps quantitatively.
Besides, it includes a few additional module (such as evomap.datasets, which provides example datasets used for these tutorials).
Example Application#
For a high-level overview of how these modules work together, we generate a market structure map for the ‘Text-Based Network Industry’ (TNIC) data, provided by Hoberg & Philips. The original data is provided at https://hobergphillips.tuck.dartmouth.edu/. If you use these data, please cite their work.
Step 1: Loading the Relationship Data#
We use a smal subsample taken from these data. The sample is included in the evomap.datasets module.
from evomap.datasets import load_tnic_sample_small
df_tnic_sample = load_tnic_sample_small()
df_tnic_sample.head()
| year | gvkey1 | gvkey2 | score | name1 | name2 | sic1 | sic2 | size1 | size2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1998 | 1078 | 1602 | 0.0274 | ABBOTT LABORATORIES | AMGEN INC | 3845 | 2836 | 74.211937 | 36.866437 |
| 1 | 1999 | 1078 | 1602 | 0.0352 | ABBOTT LABORATORIES | AMGEN INC | 3845 | 2836 | 87.854384 | 48.541222 |
| 2 | 2000 | 1078 | 1602 | 0.0348 | ABBOTT LABORATORIES | AMGEN INC | 3845 | 2836 | 70.098508 | 93.428689 |
| 3 | 2001 | 1078 | 1602 | 0.0218 | ABBOTT LABORATORIES | AMGEN INC | 3845 | 2836 | 110.299430 | 34.410965 |
| 4 | 2002 | 1078 | 1602 | 0.0366 | ABBOTT LABORATORIES | AMGEN INC | 3845 | 2836 | 40.140853 | 42.840198 |
The data consists of a time-indexed edgelist. That is, each row corresponds to a firm-pair. The ‘score’ variable captures each pair’s similarity.
To build a small subsample, we first select a handful of firms:
firms = ['APPLE INC', 'AT&T INC', 'COMCAST CORP', 'HP INC',
'INTUIT INC', 'MICROSOFT CORP', 'ORACLE CORP', 'US CELLULAR CORP',
'WESTERN DIGITAL CORP']
We then collect these firms’ pairwise relationships at a single point in time:
df_tnic_sample = df_tnic_sample.query('year == 2000').query('name1 in @firms').query('name2 in @firms')
df_tnic_sample.head()
| year | gvkey1 | gvkey2 | score | name1 | name2 | sic1 | sic2 | size1 | size2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 4796 | 2000 | 1690 | 5606 | 0.0314 | APPLE INC | HP INC | 3663 | 3570 | 60.079253 | 190.637477 |
| 4852 | 2000 | 1690 | 11399 | 0.0813 | APPLE INC | WESTERN DIGITAL CORP | 3663 | 3572 | 10.652736 | 15.988003 |
| 4884 | 2000 | 1690 | 12141 | 0.0930 | APPLE INC | MICROSOFT CORP | 3663 | 7372 | 44.120740 | 619.890226 |
| 4904 | 2000 | 1690 | 12142 | 0.0096 | APPLE INC | ORACLE CORP | 3663 | 7370 | 33.605576 | 79.457232 |
| 10644 | 2000 | 3226 | 14369 | 0.0143 | COMCAST CORP | US CELLULAR CORP | 4841 | 4812 | 40.733093 | 9.311580 |
To process these data via mapping methods, we first need to transform the edgeliste into square matrix form:
from evomap.preprocessing import edgelist2matrix
sim_mat, labels = edgelist2matrix(
df_tnic_sample, score_var = 'score', id_var_i= 'name1', id_var_j= 'name2')
sim_mat.round(2)
array([[0. , 0. , 0. , 0.03, 0. , 0.09, 0.01, 0. , 0.08],
[0. , 0. , 0. , 0. , 0. , 0. , 0. , 0.02, 0. ],
[0. , 0. , 0. , 0. , 0. , 0. , 0. , 0.01, 0. ],
[0.03, 0. , 0. , 0. , 0. , 0.06, 0.1 , 0. , 0.04],
[0. , 0. , 0. , 0. , 0. , 0.04, 0. , 0. , 0. ],
[0.09, 0. , 0. , 0.06, 0.04, 0. , 0.08, 0. , 0.06],
[0.01, 0. , 0. , 0.1 , 0. , 0.08, 0. , 0. , 0.02],
[0. , 0.02, 0.01, 0. , 0. , 0. , 0. , 0. , 0. ],
[0.08, 0. , 0. , 0.04, 0. , 0.06, 0.02, 0. , 0. ]])
As a result, we obtain a symmetric matrix of pairwise similarities.
import numpy as np
print("Smallest matrix entry: {0:.2f} \n Largest matrix entry: {1:.2f}".format(np.min(sim_mat), np.max(sim_mat)))
print("Similarity between {0} and {1}: {2:.2f}".format(labels[5], labels[6], sim_mat[5,6]))
print("Similarity between {0} and {1}: {2:.2f}".format(labels[0], labels[3], sim_mat[0,3]))
Smallest matrix entry: 0.00
Largest matrix entry: 0.10
Similarity between MICROSOFT CORP and ORACLE CORP: 0.08
Similarity between APPLE INC and HP INC: 0.03
Step 2: Preprocessing#
Different mapping methods require different input data. Here, the input data connsists of pairiwse similarities. We will map them to 2D space via Classic Multidimensional Scaling (CMDS). CMDS, however, requires pariwise distances. Among other features, evomap.preprocessing provides various transformations between such different types of relationship data.
One simple way to transform similarities to distances is by mirroring them:
from evomap.preprocessing import sim2diss
dist_mat = sim2diss(sim_mat, transformation= 'mirror')
print("Smallest matrix entry: {0:.2f} \n Largest matrix entry: {1:.2f}".format(np.min(dist_mat), np.max(dist_mat)))
print("Distance between {0} and {1}: {2:.2f}".format(labels[5], labels[6], dist_mat[5,6]))
print("Distance between {0} and {1}: {2:.2f}".format(labels[0], labels[3], dist_mat[0,3]))
Smallest matrix entry: 0.00
Largest matrix entry: 1.00
Distance between MICROSOFT CORP and ORACLE CORP: 0.92
Distance between APPLE INC and HP INC: 0.97
Step 3: Mapping relationship data to lower-dimensional space#
With all input data in the right format, you can map it to lower-dimensional space.
To do so, evomap.mapping provides implementations of multiple different mapping methods.
Here, we apply (Classic) Multidimensional Scaling (aka. Principal Coordinate Analysis):
from evomap.mapping import CMDS
model = CMDS(n_dims = 2).fit(dist_mat)
map_coords = model.Y_
The resultant model output is a 2D array of shape (n_samples, 2) containing the map coordinates.
map_coords.shape
(9, 2)
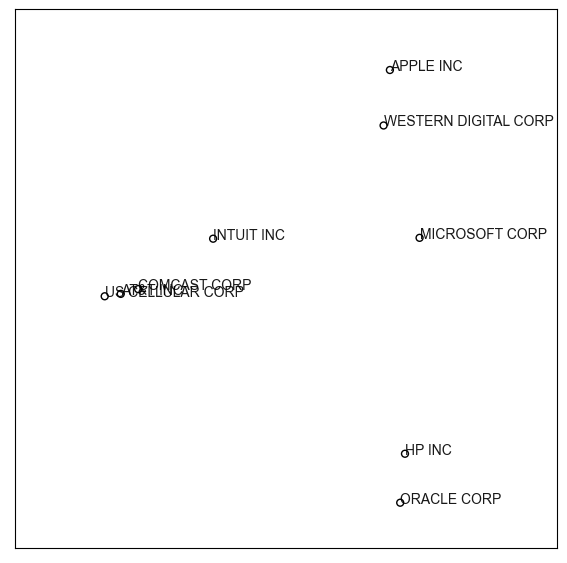
Step 4: Draw the map#
To visualize the estimated map coordinates, evomap.printer provides several functions (such as draw_map()), which can create highly customizable maps.
from evomap.printer import draw_map
draw_map(X = map_coords,
label = labels,
fig_size= (7,7))
Step 5: Evaluating maps#
Finally, evomap.metrics provides typically used metrics to evaluate the resultant maps’ goodness-of-fit (such as the hitrate of nearest neighbor recovery):
from evomap.metrics import hitrate_score
score = hitrate_score(
D = dist_mat, X = map_coords, n_neighbors = 3, input_format = 'dissimilarity')
print("Hitrate of 3-nearest neighbor recovery (adjusted or random agreement): {0:.2f}".format(score))
Hitrate of 3-nearest neighbor recovery (adjusted or random agreement): 0.63
Naturally, evomap becomes more useful when moving beyond such a very simple application.
For such more complex examples, check out the further examples.